Abstract
Background: Circular RNAs (circRNAs) constitute a class of abundant and highly conserved RNAs that show tissue- and developmental stage-specific expression. Recent studies have pointed to an important role of circRNAs in the pathology of solid tumors and differences in circRNA expression between healthy and cancerous tissue. Recently, we have shown that in the hematopoietic system, circRNA expression changes during myeloid differentiation and is also altered upon leukemic transformation.
Aims: We aimed to comprehensively characterize the circular transcriptome in healthy and leukemic hematopoietic cells and determine changes in the circRNA repertoire throughout leukemic transformation. Moreover, we studied the impact of a leukemia-specific circRNA (circBCL11B) on proliferation, differentiation and viability of leukemic cells.
Methods: We performed genome-wide circular transcriptome profiling of 16 healthy bone marrow (BM) samples and 63 AML patient samples using ribosomal RNA-depleted RNA-Seq and an in-house analysis script for the identification and quantification of circRNAs. AML patient samples included n=20 patients with a mutation of NPM1 (NPM1mut), 18 patients with splicing factor mutations (n=6 U2AF1mut, n=6 SRSF2mut, n=6 SF3B1mut) and 25 core binding factor (CBF) leukemias with t(8;21) (n=14) or inv(16) (n=11). Genes differentially expressing circRNAs were subjected to pathway analysis. Via shRNA-mediated knockdown (KD) in the OCI-AML 5 cell line, we evaluated the effect of circBCL11B on the proliferation, differentiation and viability of leukemic cells.
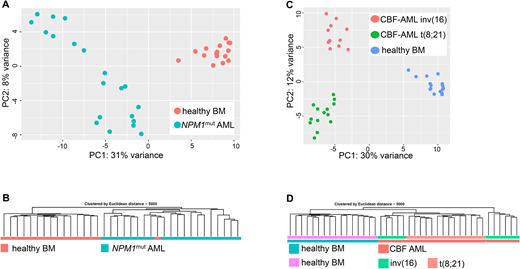
Results: Healthy BM samples could be distinguished from AML samples based on their circRNA expression profile and different AML subtypes could be discriminated (Figure 1 A-D). While for many genes differential expression distinguishing between healthy and leukemic samples was seen on both the circular and linear mRNA level, we could also identify genes with leukemia-specific circRNA deregulation irrespective of differential expression of the respective parental gene. Candidate circRNAs, like circBCL11B which was exclusively detected in AML samples, were selected for further functional evaluation. While there was no increase in apoptosis or differentiation of OCI-AML5 cells upon knockdown of circBCL11B, we observed a slight decrease in proliferation (doubling time 31h in scrambled shRNA sample vs 39h in circBCL11B KD).
Conclusions: Our work demonstrates that the circRNA repertoire and expression levels change with hematopoietic differentiation and are altered by leukemic transformation. In this "proof-of-principle" study we could identify AML-specific circRNAs and transcripts that might be involved in leukemic transformation and leukemia maintenance. The impact of spliceosome mutations on the expression of circRNAs is currently evaluated and warrants further studies. We anticipate our work to be a starting point for more comprehensive analyses of circular transcriptomes that will improve our understanding of the impact of deregulated circRNA expression on leukemogenesis.
Figure 1: Altered circular RNA expression in AML patients compared to healthy control samples. A) Principal component analysis (PCA) visualizing circRNA expression data of 16 healthy CD34+ bone marrow (BM) samples (red) and 20 AML patients with mutated NPM1 (NPM1mut) (blue). B) Unsupervised hierarchical clustering of 16 healthy BM samples (red) and 20 NPM1mut patients (blue). C) Principal component analysis (PCA) visualizing circRNA expression data of 16 healthy BM samples (blue) and 35 core binding factor (CBF) leukemias, of which 14 samples derived from patients with t(8;21) (green) and 11 samples with inv(16) (red). D) Unsupervised hierarchical clustering of 16 healthy BM samples (pink), 14 CBF leukemias with t(8;21) (light red) and 11 CBF leukemias with inv(16) (green). Expression data was generated via ribosomal RNA-depleted RNA-Seq and reads derived from circRNAs were aligned and quantified using STAR and normalized and transformed using DESeq2. PCA was performed based on 500 genes with the highest variance of circRNA expression across all samples and the unsupervised hierarchical clustering is based on 5000 circRNA-expressing genes.
Döhner:Amgen: Consultancy, Honoraria; Bristol Myers Squibb: Research Funding; Seattle Genetics: Consultancy, Honoraria; Celator: Consultancy, Honoraria; Janssen: Consultancy, Honoraria; Astellas: Consultancy, Honoraria; Jazz: Consultancy, Honoraria; Astellas: Consultancy, Honoraria; Amgen: Consultancy, Honoraria; Astex Pharmaceuticals: Consultancy, Honoraria; AbbVie: Consultancy, Honoraria; AROG Pharmaceuticals: Research Funding; Pfizer: Research Funding; Bristol Myers Squibb: Research Funding; Sunesis: Consultancy, Honoraria, Research Funding; Sunesis: Consultancy, Honoraria, Research Funding; AROG Pharmaceuticals: Research Funding; Pfizer: Research Funding; AbbVie: Consultancy, Honoraria; Novartis: Consultancy, Honoraria, Research Funding; Agios: Consultancy, Honoraria; Agios: Consultancy, Honoraria; Astex Pharmaceuticals: Consultancy, Honoraria; Janssen: Consultancy, Honoraria; Celgene: Consultancy, Honoraria, Research Funding; Jazz: Consultancy, Honoraria; Novartis: Consultancy, Honoraria, Research Funding; Seattle Genetics: Consultancy, Honoraria; Celator: Consultancy, Honoraria; Celgene: Consultancy, Honoraria, Research Funding. Bullinger:Bayer Oncology: Research Funding; Janssen: Speakers Bureau; Sanofi: Research Funding, Speakers Bureau; Jazz Pharmaceuticals: Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Amgen: Honoraria, Speakers Bureau; Pfizer: Speakers Bureau; Bristol-Myers Squibb: Speakers Bureau; Novartis: Honoraria, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal